Abstract
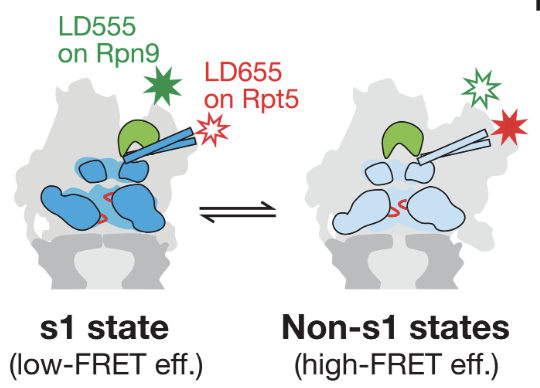
The 26S proteasome is the principal macromolecular machine responsible for protein degradation in eukaryotes. However, little is known about the detailed kinetics and coordination of the underlying substrate-processing steps of the proteasome, and their correlation with observed conformational states. Here, we used reconstituted 26S proteasomes with unnatural amino-acid-attached fluorophores in a series of FRET- and anisotropy-based assays to probe substrate-proteasome interactions, the individual steps of the processing pathway, and the conformational state of the proteasome itself. We develop a complete kinetic picture of proteasomal degradation, which reveals that the engagement steps prior to substrate commitment are fast relative to subsequent deubiquitination, translocation, and unfolding. Furthermore, we find that non-ideal substrates are rapidly rejected by the proteasome, which thus employs a kinetic proofreading mechanism to ensure degradation fidelity and substrate prioritization.
*These authors contributed equally to this work.